RStudio-JupyterHPC
We offer the possibility of running RStudio instances on the interactive partitions of our HPC clusters, through the JupyterHPC container platform. The advantages of this approach include more flexible resource allocation, access to your usual files in your home folder, and the possibility of rapidly creating new RStudio containers tailored to your specific requirements.
For calculations that need compute resources over a long duration, please submit a batch job to the appropriate Slurm partition instead of using the RStudio instance.
Starting your RStudio-JupyterHPC instance
Project portal, SCC and NHR users: https://jupyter.hpc.gwdg.de
- Go to https://jupyter.hpc.gwdg.de and log in with your usual account.
- “Start my server”, if the button appears.
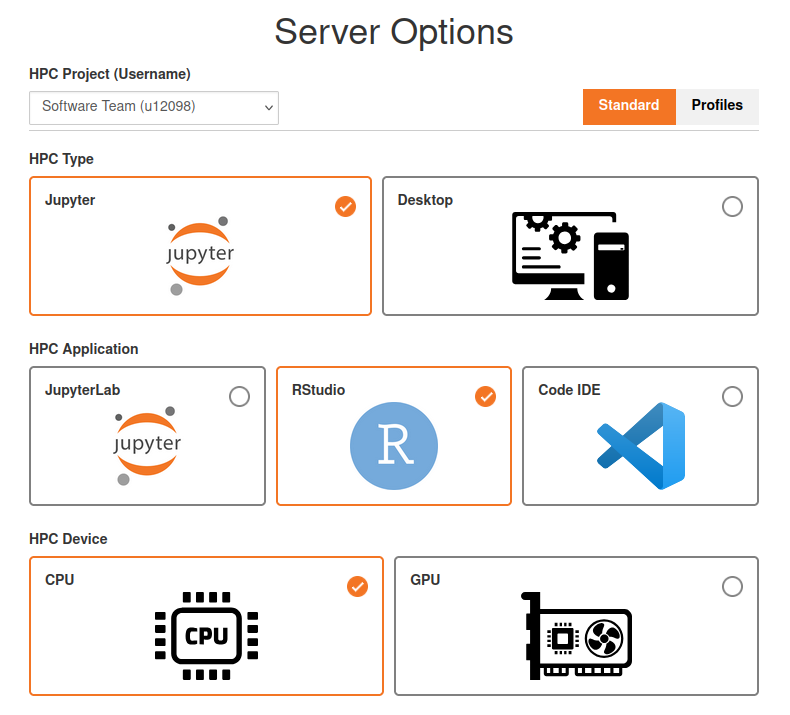
- Select the appropriate entry from the “HPC Project (Username)” dropdown. Select the Project Portal username (u12345) corresponding to your project.
- Select the “Jupyter” and then “RStudio” cards, and “CPU” as the HPC Device.
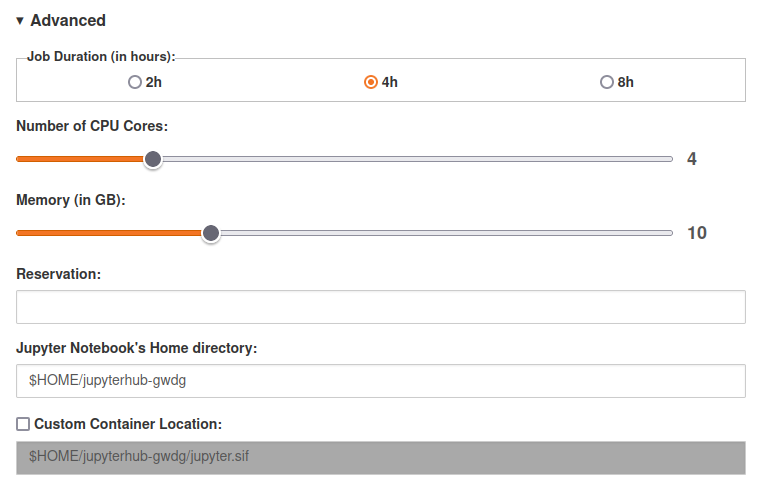
- Under Advanced, set a reasonable amount of resources to use, the defaults might be way too high for simple R jobs! Reasonable defaults for simple jobs are: 1-2 CPUs, 1-2 GBs of RAM/Memory, max. 8 hours runtime. If you know that you will need it, you can request more CPUs or RAM/Memory.
In the end, your configuration should look similar to:
These containers start up as jobs in an interactive cluster partition, and so will expire after the time given in the initial options (as will any R jobs that you have left running). The maximum allowed time is currently 8 hours. If your jobs require longer running times, please let us know.
Click on start server, spawning might take 1-2 minutes (if the server fails to start, it will time out on its own! Don’t refresh the page!)
Once your server starts, you will be directly logged into an RStudio session.
If you experience problems starting your server, please provide any errors shown by your browser, as well as the contents of
~/current.jupyterhub.notebook.log(don’t start another notebook or it will overwrite this file!). A common cause of failure to spawn is running out of disk space quota, so please first check that you still have space for new files!
Stopping your server/notebook/RStudio instance
- In RStudio, press the red button up and to the right. (Other equivalent buttons under File and Session do not work for the time being)
- This should return you to the Jupyter GUI. Click on Stop My Server, you might need to wait a minute or two.
Of course you can also just let your session expire after the previously given runtime.
Accessing data from your legacy account
Data from the old RStudio instance went directly to your legacy user’s home folder. This home folder is not directly available from the new RStudio instance which uses queues on the Emmy and Grete islands (see HOME folder documentation for more information on this). There are two options here:
Option 1: If you do not have too many files or too large files, the best option is to copy over data from your legacy account to your project account. See the Data Migration Guide for details on how to do this. In short, you will have to:
- Check that your legacy user and your new project user share a group (which they should if everything is set up correctly).
- Use
chgrpandchmodon the folders of your legacy account so they can be accessed by other members of your group (in this case, your project account) - Move or copy the folders from the legacy home to the project account folder.
- This needs to be done while logged in to the SCC cluster (where both your legacy home and your project home are available ) via SSH.
Option 2: If you have too many files to transfer everything to the quota of your project user, you can work by going through SCRATCH:
On your legacy account, on the SCC:
- Copy your relevant data to
/scratch-scc/users/USERNAME. Data inscratchdoes not count against your disk quota. - Please take into account that
scratchshould be used for non-permanent data only. Only use scratch to store data that you can easily recreate. You should always keep a copy of final results and input files that are difficult to reconstruct in your home folders. - Make the relevant changes to the permissions of your
scratchfolder, as explained in Data Migration Guide and Option 1.
- Copy your relevant data to
On your RStudio instance:
- You can now access the data at
/scratch-scc/users/USERNAME. - Tell R to process and load your data from
scratch, and output your results to your project user home folder. - If you know how to work with soft/symbolic links, you can create one to the
scratchlocation for convenience. - Once again, only use
scratchfor temporary files and data! Anything that is difficult to reconstruct should live in your home folders!
- You can now access the data at
Warning
Access to the data in the home folders of legacy accounts will be slowly phased out as time goes on, and become more difficult to access. Dedicated transfer nodes will be provided, but will make accessing your old data more complicated. Please consider Options 1 and 2 as temporary solutions to help you in the transition. You should fully migrate your work to a project user as soon as possible!
Installing packages
The RStudio containers already contain a large number of the more commonly requested R packages. If you need other packages, install them the usual way from inside the RStudio instance in the container. They will be installed to your home folder, and be available whenever you restart the container.
Newer R version
If you require a newer R version for your RStudio instance due to some specific packages, let us know and we can build an updated container for you.
Retrieving your old RStudio packages
NOTE: This will end up installing A LOT of packages, since it will also reinstall any packages that might be slightly newer than the ones already available in the container. I recommend picking only those libraries you actually work with instead of this brute-force approach.
- On your old or personal RStudio instance, go to the R tab and:
ip <- installed.packages()[,1]
write(ip,"rpackages_in_4.2.0.txt")- Copy the created file to the cluster corresponding to your account. In the new RStudio instance now do:
ip <- readLines("rpackages_in_4.2.0.txt")
install.packages(ip)- Some packages might have been installed through the R package BiocManager, in which case:
ip <- readLines("rpackages_in_4.2.0.txt")
BiocManager::install(ip)More information on JupyterHPC and Containers
Creating containers for JupyterHPC , with a couple of example container definition files.
Using apptainer (to create and test new containers). Notice you need to run apptainer from inside a Slurm job! Ideally use an interactive job in an interactive queue for this purpose.
Advanced: Testing the RStudio container & using it for Slurm jobs
If you want to test the environment of the RStudio container without the burden/extra environment of Jupyter and RStudio, you can run the container directly. You can also use this approach to start up and use the container in batch (that is, non-interactive) mode.
- Start an interactive (or batch) job.
- Load the apptainer module.
apptainer run container.sifwill “log into” the container.
You can also build your own container from the examples in the JupyterHPC page, or following this recipe/.def file used for the RStudio containers (might be out of date! No guarantees it will work correctly). Recipe might take about an hour to build and the resulting container file will be a couple of GBs large:
CLICK ME for a large container definition file
Bootstrap: docker
#From: condaforge/miniforge3
From: ubuntu:jammy
%post
export DEBIAN_FRONTEND=noninteractive
apt update
apt upgrade -y
# Install Julia
# Not available in 22.04 repos
# apt install -y julia
# echo 'ENV["HTTP_PROXY"] = "http://www-cache.gwdg.de:3128"' >> /etc/julia/startup.jl
# echo 'ENV["HTTPS_PROXY"] = "http://www-cache.gwdg.de:3128"' >> /etc/julia/startup.jl
##################
# R and packages #
##################
apt install -y --no-install-recommends software-properties-common dirmngr
apt install -y wget curl libcurl4-openssl-dev git-all
wget -qO- https://cloud.r-project.org/bin/linux/ubuntu/marutter_pubkey.asc | tee -a /etc/apt/trusted.gpg.d/cran_ubuntu_key.asc
add-apt-repository "deb https://cloud.r-project.org/bin/linux/ubuntu $(lsb_release -cs)-cran40/"
#add-apt-repository ppa:c2d4u.team/c2d4u4.0+
apt update
apt install -y \
r-base \
r-base-dev \
r-cran-caret \
r-cran-crayon \
r-cran-devtools \
r-cran-forecast \
r-cran-hexbin \
r-cran-htmltools \
r-cran-htmlwidgets \
r-cran-plyr \
r-cran-randomforest \
r-cran-rcurl \
r-cran-reshape2 \
r-cran-rmarkdown \
r-cran-rodbc \
r-cran-rsqlite \
r-cran-shiny \
r-cran-tidyverse \
r-cran-rcpp \
libfftw3-3 libfftw3-dev libgdal-dev
apt install -y \
r-bioc-annotationdbi r-cran-bh r-bioc-biobase r-bioc-biocfilecache r-bioc-biocgenerics r-bioc-biocio \
r-cran-biocmanager r-cran-biocmanager r-bioc-biocneighbors r-bioc-biocparallel r-bioc-biocsingular r-bioc-biocversion \
r-bioc-biostrings r-cran-cairo r-bioc-complexheatmap r-cran-dbi r-cran-ddrtree r-bioc-deseq2 \
r-bioc-delayedarray r-bioc-delayedmatrixstats r-cran-fnn r-cran-formula r-bioc-geoquery r-bioc-go.db \
r-bioc-gosemsim r-bioc-genomeinfodb r-bioc-genomeinfodbdata r-bioc-genomicalignments r-bioc-genomicfeatures r-bioc-genomicranges \
r-cran-getoptlong r-cran-globaloptions r-bioc-hdf5array r-bioc-hsmmsinglecell r-cran-hmisc r-bioc-iranges \
r-bioc-keggrest r-cran-kernsmooth r-cran-mass r-cran-matrix r-bioc-matrixgenerics r-cran-nmf \
r-cran-r.cache r-cran-r.methodss3 r-cran-r.oo r-cran-r.utils r-cran-r6 r-cran-rann \
r-bioc-rbgl r-cran-rcolorbrewer r-cran-rcurl r-cran-rmysql r-cran-rocr r-cran-rsqlite \
r-cran-rspectra r-cran-runit r-cran-rcpp r-cran-rcppannoy r-cran-rcpparmadillo r-cran-rcppeigen \
r-cran-rcpphnsw r-cran-rcppparallel r-cran-rcppprogress r-cran-rcpptoml r-bioc-residualmatrix r-bioc-rhdf5lib \
r-bioc-rhtslib r-bioc-rsamtools r-cran-rserve r-cran-rtsne r-bioc-s4vectors r-bioc-scaledmatrix \
r-cran-seurat r-cran-seuratobject r-bioc-singlecellexperiment r-cran-sparsem r-cran-stanheaders r-bioc-summarizedexperiment \
r-cran-v8 r-cran-vgam r-cran-venndiagram r-cran-xml r-bioc-xvector r-cran-abind \
r-cran-acepack r-bioc-affxparser r-bioc-affy r-bioc-affyio r-bioc-annotate r-cran-ape \
r-cran-askpass r-cran-assertthat r-cran-backports r-cran-base64enc r-bioc-beachmat r-cran-beeswarm \
r-cran-bibtex r-cran-bindr r-cran-bindrcpp r-bioc-biocviews r-bioc-biomart r-cran-bit \
r-cran-bit64 r-cran-bitops r-cran-blob r-bioc-bluster r-cran-boot r-cran-brew \
r-cran-brio r-cran-broom r-cran-catools r-cran-cachem r-cran-callr r-cran-car \
r-cran-cardata r-cran-cellranger r-cran-checkmate r-cran-circlize r-cran-class r-cran-classint \
r-cran-cli r-cran-clipr r-cran-clue r-cran-cluster r-cran-coda r-cran-codetools \
r-cran-colorspace r-cran-combinat r-cran-commonmark r-cran-corpcor r-cran-corrplot r-cran-covr \
r-cran-cowplot r-cran-cpp11 r-cran-crayon r-cran-credentials r-cran-crosstalk r-cran-curl \
r-cran-data.table r-cran-dbplyr r-cran-deldir r-cran-desc r-cran-devtools r-cran-dichromat \
r-cran-diffobj r-cran-digest r-cran-doparallel r-cran-dorng r-cran-docopt r-cran-downlit \
r-cran-downloader r-cran-dplyr r-cran-dqrng r-cran-dtplyr r-bioc-edger r-cran-ellipse \
r-cran-ellipsis r-cran-evaluate r-cran-expm r-cran-fansi r-cran-farver r-cran-fastica \
r-cran-fastmap r-cran-fastmatch r-cran-ff r-cran-fitdistrplus r-cran-flexmix r-cran-forcats \
r-cran-foreach r-cran-foreign r-cran-formatr r-cran-fs r-cran-furrr r-cran-futile.logger \
r-cran-futile.options r-cran-future r-cran-future.apply r-cran-gargle r-cran-gdata r-bioc-genefilter \
r-bioc-geneplotter r-cran-generics r-cran-gert r-cran-getopt r-cran-ggalluvial r-cran-ggbeeswarm \
r-cran-ggforce r-cran-ggplot2 r-cran-ggpubr r-cran-ggraph r-cran-ggrepel r-cran-ggridges \
r-cran-ggsci r-cran-ggsignif r-cran-gh r-cran-gitcreds r-bioc-glmgampoi r-cran-globals \
r-cran-glue r-cran-goftest r-cran-googledrive r-cran-googlesheets4 r-cran-gplots r-bioc-graph \
r-cran-graphlayouts r-cran-gridbase r-cran-gridextra r-cran-gridgraphics r-cran-gtable r-cran-gtools \
r-cran-haven r-cran-hdf5r r-cran-here r-cran-highr r-cran-hms r-cran-htmltable \
r-cran-htmltools r-cran-htmlwidgets r-cran-httpuv r-cran-httr r-cran-ica r-cran-ids \
r-cran-igraph r-cran-ini r-cran-inline r-cran-irlba r-cran-isoband r-cran-iterators \
r-cran-itertools r-cran-jpeg r-cran-jquerylib r-cran-jsonlite r-cran-knitr r-cran-labeling \
r-cran-lambda.r r-cran-later r-cran-lattice r-cran-latticeextra r-cran-lazyeval r-cran-leiden \
r-cran-lifecycle r-bioc-limma r-cran-listenv r-cran-lme4 r-cran-lmtest r-cran-locfit \
r-cran-loo r-cran-lubridate r-cran-magrittr r-bioc-makecdfenv r-cran-markdown r-cran-matrixstats \
r-cran-mclust r-cran-memoise r-bioc-metapod r-cran-mgcv r-cran-mime r-cran-miniui \
r-cran-minqa r-cran-mnormt r-cran-modelr r-cran-modeltools r-bioc-monocle r-bioc-multtest \
r-cran-munsell r-cran-network r-cran-nleqslv r-cran-nlme r-cran-nloptr r-cran-nnet \
r-cran-numderiv r-bioc-oligo r-bioc-oligoclasses r-cran-openssl r-cran-pander r-cran-parallelly \
r-cran-patchwork r-cran-pbapply r-cran-pbkrtest r-cran-pbmcapply r-bioc-pcamethods r-cran-pheatmap \
r-cran-pillar r-cran-pkgbuild r-cran-pkgconfig r-cran-pkgload r-cran-pkgmaker r-cran-plogr \
r-cran-plotly r-cran-plyr r-cran-png r-cran-polyclip r-cran-polynom r-cran-praise \
r-bioc-preprocesscore r-cran-prettyunits r-cran-processx r-cran-progress r-cran-progressr r-cran-promises \
r-cran-proto r-cran-proxy r-cran-ps r-cran-pscl r-cran-psych r-cran-purrr \
r-cran-qlcmatrix r-cran-quadprog r-cran-quantreg r-bioc-qvalue r-cran-ragg r-cran-randomforest \
r-cran-rappdirs r-cran-raster r-cran-rcmdcheck r-cran-readr r-cran-readxl r-cran-registry \
r-cran-rematch r-cran-rematch2 r-cran-remotes r-cran-reprex r-cran-reshape r-cran-reshape2 \
r-cran-restfulr r-cran-reticulate r-cran-rex r-bioc-rhdf5 r-bioc-rhdf5filters r-cran-rjags \
r-cran-rjson r-cran-rlang r-cran-rmarkdown r-cran-rngtools r-cran-roxygen2 r-cran-rpart \
r-cran-rprojroot r-cran-rsample r-cran-rstan r-cran-rstatix r-cran-rstudioapi r-cran-rsvd \
r-bioc-rtracklayer r-cran-rversions r-cran-rvest r-cran-s2 r-cran-sandwich r-cran-sass \
r-cran-scales r-bioc-scater r-cran-scattermore r-bioc-scran r-cran-sctransform r-bioc-scuttle \
r-cran-selectr r-cran-sessioninfo r-cran-sf r-cran-sfsmisc r-cran-shape r-cran-shiny \
r-cran-sitmo r-cran-slam r-cran-slider r-cran-sna r-cran-snow r-cran-sourcetools \
r-cran-sp r-cran-spdata r-bioc-sparsematrixstats r-cran-sparsesvd r-cran-spatial r-cran-spatstat.data \
r-cran-spatstat.geom r-cran-spatstat.random r-cran-spatstat.sparse r-cran-spatstat.utils r-cran-spdep r-cran-statmod \
r-cran-statnet.common r-cran-stringi r-cran-stringr r-cran-survival r-bioc-sva r-cran-svglite \
r-cran-sys r-cran-systemfonts r-cran-tensor r-cran-terra r-cran-testthat r-cran-textshaping \
r-cran-tibble r-cran-tidygraph r-cran-tidyr r-cran-tidyselect r-cran-tidyverse r-cran-timedate \
r-cran-timeseries r-cran-tinytex r-cran-tweenr r-cran-tzdb r-cran-udunits2 r-cran-units \
r-cran-usethis r-cran-utf8 r-cran-uuid r-cran-uwot r-cran-vctrs r-cran-vipor \
r-cran-viridis r-cran-viridislite r-cran-vroom r-cran-waldo r-cran-warp r-cran-webshot \
r-cran-whisker r-cran-withr r-cran-wk r-cran-xfun r-cran-xml2 r-cran-xopen \
r-cran-xtable r-cran-yaml r-cran-zeallot r-cran-zip r-bioc-zlibbioc r-cran-zoo
# rcpp: solves some issues with -Wformat errors when installing various packages under R4.4, the package manager version of RCpp is not new enough
echo ""
echo "#######################################"
echo "# Starting installation of R packages #"
echo "#######################################"
echo ""
echo 'install.packages("Rcpp")' >> packages.R
echo 'BiocManager::install(version = "3.19", update=FALSE, ask=FALSE)' >> packages.R
echo 'ip <-c("eseis","CellChat","ClusterProfiler","RCppML","SeuratData","SeuratDisk","SeuratWrappers","fgsea")' >> packages.R
echo 'install.packages(ip, Ncpus=4)' >> packages.R
echo 'BiocManager::install(ip, update=FALSE, ask=FALSE)' >> packages.R
# Run installation and divert output to dev/null, there is A LOT of output
# Comment this out if you are just testing stuff cos it is going to take a while
Rscript packages.R 2>&1 >/dev/null
rm packages.R
echo ""
echo "########################################"
echo "# Done with installation of R packages #"
echo "########################################"
echo ""
###########
# RStudio #
###########
apt install -y libclang-dev lsb-release psmisc sudo libssl-dev
ubuntu_release=$(lsb_release --codename --short)
wget https://download2.rstudio.org/server/${ubuntu_release}/amd64/rstudio-server-2023.12.1-402-amd64.deb
dpkg --install rstudio-server-2023.12.1-402-amd64.deb
rm rstudio-server-2023.12.1-402-amd64.deb
echo 'ftp_proxy=http://www-cache.gwdg.de:3128' >> /usr/lib/R/etc/Renviron.site
echo 'https_proxy=http://www-cache.gwdg.de:3128' >> /usr/lib/R/etc/Renviron.site
echo 'http_proxy=http://www-cache.gwdg.de:3128' >> /usr/lib/R/etc/Renviron.site
echo '' >> /usr/lib/R/etc/Renviron.site
# Other stuff
apt install -y vim
apt install -y default-jre # required for gipptools
#########
# Conda #
#########
# Among other stuff, installs Jupyter env.
# Install miniconda to /miniconda
condash="Miniconda3-py310_24.5.0-0-Linux-x86_64.sh"
curl -LO "http://repo.continuum.io/miniconda/${condash}"
bash ${condash} -p /opt/conda -b
rm ${condash}
PATH=/opt/conda/bin:${PATH}
conda update -y conda
conda init
conda install --quiet --yes -c conda-forge \
'ipyparallel' \
'jupyter-rsession-proxy' \
'notebook' \
'jupyterhub==2.3.1' \
'jupyterlab'
conda install --quiet --yes -c conda-forge \
dgl \
igraph \
keras \
pandas \
pydot \
scikit-learn \
scipy \
seaborn
%environment
# required so JupyterHPC can find jupyterhub-singleuser
export PATH=$PATH:/opt/conda/binTroubleshooting & FAQ
- Please adjust your utilized resources to reasonable numbers, since you will be sharing the interactive partition nodes with others (1-2 CPUs, 1-2 GBs of RAM/Memory, max. 8 hours runtime).
- Your usual home folder files should be accessible from the container.
- You can install packages as usual with install.packages, or if you think it will be a popular package, request a centralized installation.
- If you are experiencing strange issues, check you do not have leftover configuration files from other RStudio instances, e.g. ~/.R/Makevars or an old .RData file, in your home folder.
- External modules (
module load) are NOT accessible.
Known Issues
- $HOME might not be set up correctly in the Terminal tab (it is correct from the R tab in RStudio), so you might want to change it if some scripts of yours depend on this. This on RStudio’s Terminal tab might fix it:
export HOME=/usr/users/$USER- You can also ignore any LC_whatever error messages related to locale configuration.
- Function help with F1 might show a “Firefox can’t open embedded page” error.