SpiNNaker
Introduction
SpiNNaker is the neuromorphic computing architecture from SpiNNcloud. The SpiNNaker architecture has been in development for a couple of decades, with different iterations. We currently host the recently developed SpiNNaker-2 version, and will eventually provide access to 4 boards.
SpiNNaker can be used to run typical spiking neuron neuromorphic simulations. This includes simulations of nervous tissue, AI/ML algorithms, graph, constraint, network and optimization problems, signal processing, and control loops. But, additionally, due to its flexible ARM based architecture, it can be made to run other non-neuromorphic simulations (for example, solving distributed systems of partial differential equations).
Finally, due to its sparse and efficient message passing architecture and efficient ARM-based cores, the hardware exhibits low power consumption when compared with similar more traditional configurations and algorithms.
For more information and reference material, please consult this article in the January/February 2024 issue of GWDG News.

SpiNNaker boards
Architecture
The hardware architecture of SpiNNaker-2 is naturally quite complex, but it can be reduced to two main components relevant to the average end user:
- An efficient message passing network with various network-on-a-chip level components. This is vital for efficient communication and routing between neurons that might reside in different chips or even different boards. Messages can even account for delay incurred during message transmission.
- A large number of low-powered but efficient ARM cores, organized in a hierarchical, tiered configuration. It is important to understand this setup, because it can affect the structure and efficiency of your network. Each core has a limited number of neurons and synapses that it can fit (this number changes as models become more complex and require more memory and compute cycles). Naturally, communication is more efficient within a core than across cores in different boards. The following image shows the distribution of cores up to the multi-board level. Pay particular attention to the PE and QPE terminology ([Quad] Processing Element).
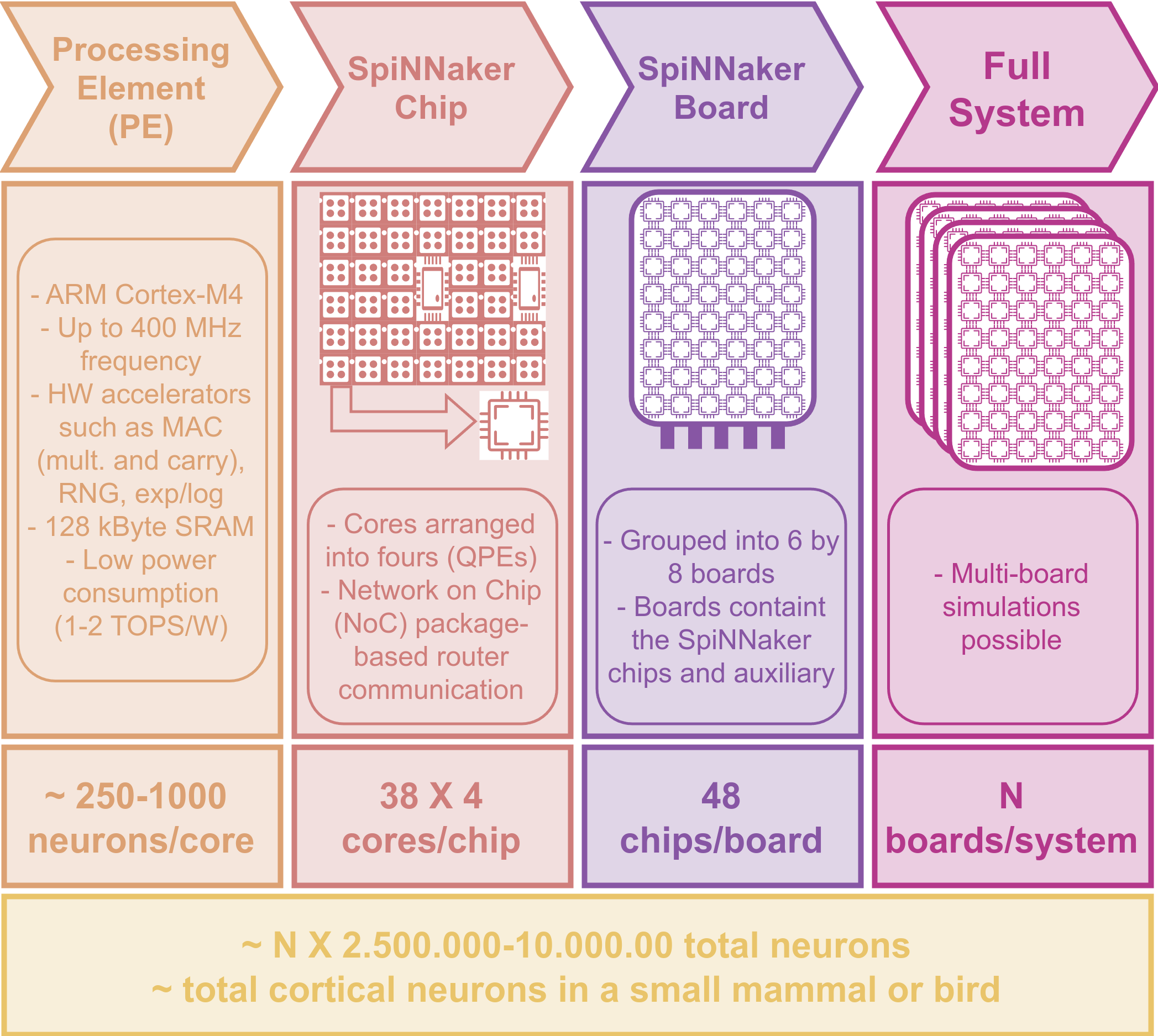
SpiNNaker core architecture
How to Access
Access to SpiNNaker is currently possible through our Future Technology Platform (FTP). For this an account is required (usual GWDG account, or AcademicCloudID for external users), that then needs to be explicitly enabled to access the FTP. For more information, check our documentation on getting an account.
Access requests currently run through KISSKI (researchers and companies) and NHR (researchers at Universities only). Please consult their documentation and eventually request a project to test and utilize SpiNNaker. We also highly recommend you try to run some neuromorphic simulations with PyNN or other neuromorphic libraries first. If you have questions, you can also reach out through one of our support channels .
How to Use
The easiest way of approaching SpiNNaker simulations at the moment is through its Python library. This library is available through an open repository from SpiNNcloud. It is also provided on the FTP as an Apptainer container, which also includes the necessary device compilers targeting the hardware. The py-spinnaker2 library syntax is very similar to PyNN’s (and a PyNN integration is planned). For this reason, we recommend learning how to set up simulations with PyNN first, and testing your algorithms on PyNN plus a regular CPU simulator such as Neuron or NEST. See our page on neuromorphic tools for more information.
Container
The container is available at /sw/containers/spinnaker, from the FTP system only. You can start it up and use it as any other container. See the page for Apptainer for more information. Relevant libraries are to be found in /usr/spinnaker from inside the container. The container also includes some neuromorphic libraries such as PyNN, for convenience. Others can be included upon request. See also the neuromorphic tools container if you want to test some non-SpiNNaker libraries.
How a SpiNNaker simulation works
SpiNNaker functions as a heterogeneous accelerator. Working with this is in concept similar to a GPU, where the device has its own memory address space, and work proceeds in a batched approach. A simulation will look roughly like this:
- Set up your neurons, networks, models, and any variables to be recorded.
- Issue a Python command that submits your code to the device.
- Your code gets compiled and uploaded to the board.
- Board runs the simulation.
- Once the simulation ends, you can retrieve the memory stored on the board.
- You can continue working with the results returned by the board.
Notice that you don’t have direct control of the board while it is running the simulation (although I believe some callback functions might be available).
Communicating with the board
Currently, the hardware can be accessed from the log-in node of the FTP. This will be subject to change as our installation of the SpiNNaker hardware and the FTP itself evolve, and the boards will then only be accessible from proper, dedicated compute nodes that require going through a scheduler.
Tip
At present, you need to know the address of the board to communicate with it. This will be provided once you gain access to the FTP system.
py-spinnaker2: Code and Examples
You can find the py-spinnaker2 folder in /usr/spinnaker/py-spinnaker2 in the container. Examples are provided in the examples folder. Particularly useful are the folders snn and snn_profiling. Python code for the higher level routines available in /src.
Example 1: A minimal network
This example, taken from the basic network examples in the py-spinnaker2 repository, sets up a network with just 3 neurons. 2 input “neurons” that are just a spike source (they emit a spike at fixed, given times), one an excitatory and the other one an inhibitory pulse, and then a single LIF (leaky integrate and fire) neuron, whose voltage we track.
from spinnaker2 import hardware, snn
# Input Population
## create stimulus population with 2 spike sources
input_spikes = {0: [1, 4, 9, 11], 1: [20, 30] } # will spike at the given times
stim = snn.Population(size=2, neuron_model="spike_list", params=input_spikes, name="stim")
# Core Population
## create LIF population with 1 neuron
neuron_params = {
"threshold": 10.0,
"alpha_decay": 0.9,
"i_offset": 0.0,
"v_reset": 0.0,
"reset": "reset_by_subtraction",
}
pop1 = snn.Population(size=1, neuron_model="lif",
params=neuron_params,
name="pop1", record=["spikes", "v"])
# Projection: Connect both populations
## each connection has 4 entries: [pre_index, post_index, weight, delay]
## for connections to a `lif` population:
## - weight: integer in range [-15, 15]
## - delay: integer in range [0, 7]. Actual delay on the hardware is: delay+1
conns = []
conns.append([0, 0, 4, 1]) # excitatory synapse with weight 4 and delay 1
conns.append([1, 0, -3, 2]) # inhibitory synapse with weight -3 and delay 1
proj = snn.Projection(pre=stim, post=pop1, connections=conns)
# Network
## create a network and add population and projections
net = snn.Network("my network")
net.add(stim, pop1, proj)
# Hardware
## select hardware and run network
hw = hardware.SpiNNaker2Chip(eth_ip="boardIPaddress")
timesteps = 50
hw.run(net, timesteps)
# Results
## get_spikes() returns a dictionary with:
## - keys: neuron indices
## - values: lists of spike times per neurons
spike_times = pop1.get_spikes()
## get_voltages() returns a dictionary with:
## - keys: neuron indices
## - values: numpy arrays with 1 float value per timestep per neuron
voltages = pop1.get_voltages()The resulting LIF neuron voltage dynamic looks like:
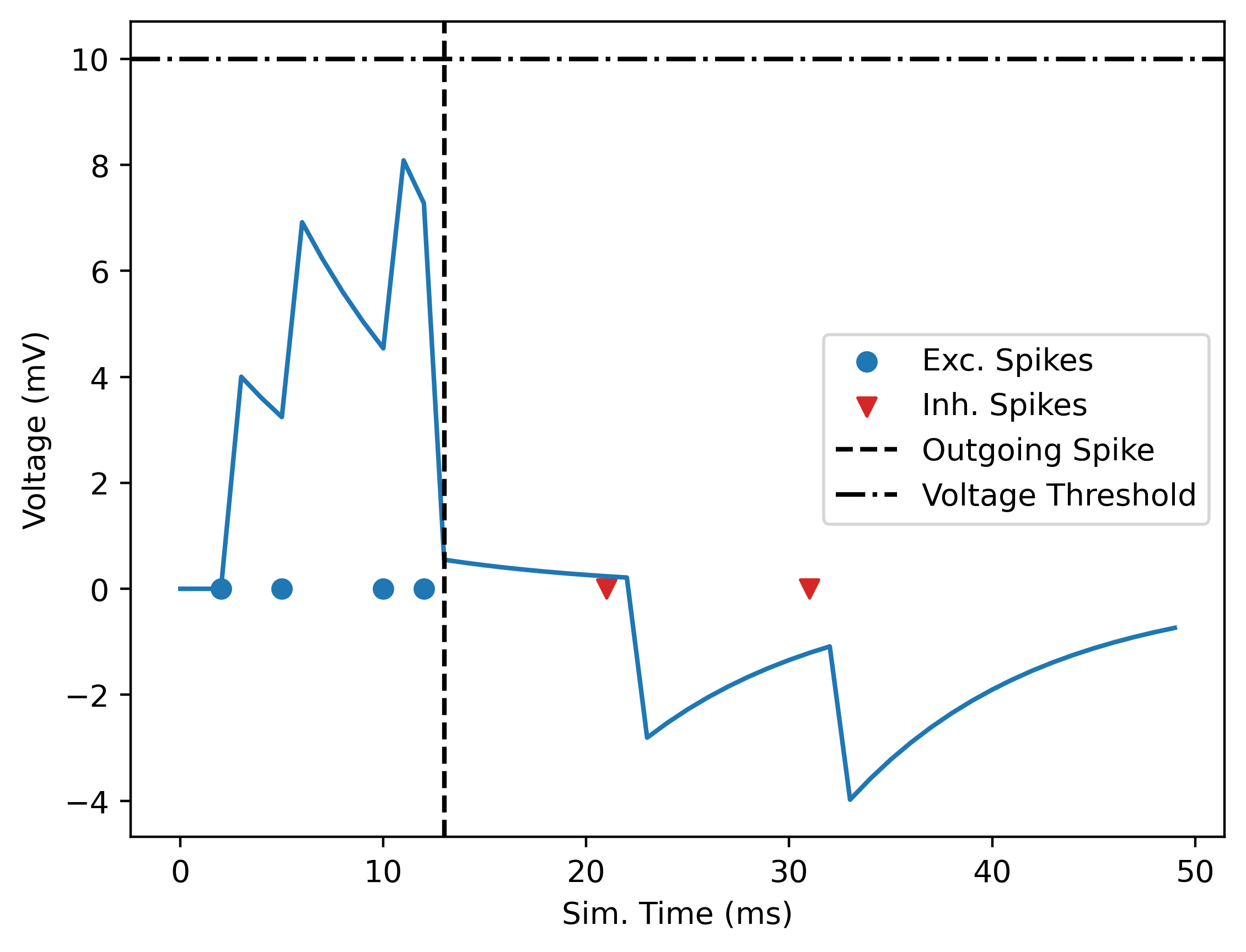
Neuron voltage
Running this code results in quite a bit of SpiNNaker specific output, which can tell you some details about your simulation. This is the output in this case, with some comments:
Used 2 out of 152 cores # 152 PEs per board
Sim duration: 0.05 s # As requested, 50 timesteps at 1 ms each
Actual duration (incl. offset): 0.060000000000000005
INFO: open and bind UDP socket
INFO: Running test ...
configure hardware
There are 1QPEs
Enabled QPEs.
INFO: start random bus...
Enabled random bus
# Loading program into (Q)PEs
INFO: QPE (1,1) PE 0): loading memory from file /s2-sim2lab-app/chip/app-pe/s2app/input_spikes_with_routing/binaries/s2app_arm.mem...
INFO: QPE (1,1) PE 1): loading memory from file /s2-sim2lab-app/chip/app-pe/s2app/lif_neuron/binaries/s2app_arm.mem...
Loaded mem-files
Data written to SRAM
PEs started
Sending regfile interrupts for synchronous start
Run experiment for 0.06 seconds
Experiment done
# Reading back results from board's memory
Going to read 4000 from address 0xf049b00c (key: PE_36_log)
Going to read 4000 from address 0xf04bb00c (key: PE_37_log)
Going to read 500 from address 0xf04b00d0 (key: PE_37_spike_record)
Going to read 100 from address 0xf04b08a0 (key: PE_37_voltage_record)
Results read
interface free again
Debug size: 48
n_entries: 12
Log PE(1, 1, 0)
magic = ad130ad6, version = 1.0
sr addr:36976
Debug size: 604
n_entries: 151
Log PE(1, 1, 1)
magic = ad130ad6, version = 1.0
pop_table_info addr: 0x8838
pop_table_info value: 0x10080
pop_table addr: 0x10080
pop_table_info.address: 0x10090
pop_table_info.length: 2
pop table address 0:
pop table address 1:
pop table address 2:
== Population table (2 entries) at address 0x10090 ==
Entry 0: key=0, mask=0xffffffff, address=0x100b0, row_length=4
Entry 1: key=1, mask=0xffffffff, address=0x100c0, row_length=4
global params addr: 0x10060
n_used_neurons: 1
record_spikes: 1
record_v: 1
record_time_done: 0
profiling: 0
reset_by_subtraction: 1
spike_record_addr: 0x8854
Neuron 0 spiked at time 13
Read spike record
read_spikes(n_neurons=1, time_steps=50, max_atoms=250)
Read voltage record
read_voltages(n_neurons=1, time_steps=50)
Duration recording: 0.0030603408813476562
{0: [13]}
{0: array([ 0. , 0. , 0. , 4. , 3.6 ,
3.2399998 , 6.916 , 6.2243996 , 5.601959 , 5.0417633 ,
(...)
-1.1239064 , -1.0115157 , -0.91036415, -0.8193277 , -0.7373949 ],
dtype=float32)}Tip
Notice that the simulation also generates a pair of human-readable files in JSON format (spec.json and results.json). These contain the data retrieved from the SpiNNaker board, including for example neuron voltages, and can be used for post-processing, without needing to rerun the whole calculation.
Example 2: Testing limits
The SpiNNaker library has a number of Exception errors that you can catch. The following example shows how to test the maximum number of synapses/connections between 2 populations, that can be fit in SpiNNaker:
from spinnaker2 import hardware, snn
def run_model(pops=2, nneurons=50, timesteps=0):
# Create population
neuron_params = {
"threshold": 100.0,
"alpha_decay": 0.9,
}
pops_and_projs=[]
for ipop in range(pops):
pop=snn.Population(size=nneurons, neuron_model="lif", params=neuron_params, name="pop{}".format(ipop))
# Limit the number of neurons of this population in a given PE/core
# pop.set_max_atoms_per_core(50) #tradeoff between neurons per core and synapses per population
pops_and_projs.append(pop)
# Set up synpases
w = 2.0 # weight
d = 1 # delay
## All to all connections
conns = []
for i in range(nneurons):
for j in range(nneurons):
conns.append([i, j, w, d])
for ipop in range(pops-1):
proj=snn.Projection(pre=pops_and_projs[ipop], post=pops_and_projs[ipop+1], connections=conns)
pops_and_projs.append(proj) #adding the projections to the pop list too for convenience
# Put everything into network
net = snn.Network("my network")
net.add(*pops_and_projs) #unpack list
# Dry run with mapping_only set to True, we don't even need board IP
hw = hardware.SpiNNaker2Chip()
hw.run(net, timesteps, mapping_only=True)
def sweep(pops=2, nneurons=[1, 10, 50, 100, 140, 145, 150, 200, 250]):
# max number of pre neurons with 250 post neurons
timesteps = 0
best_nneurons = 0
for nn in nneurons:
try:
run_model(pops=pops, nneurons=nn, timesteps=timesteps)
best_nneurons = nn
except MemoryError:
print(f"Could not map network with {nn} neurons and {pops} populations")
break
max_synapses = best_nneurons**2
max_total_synapses = max_synapses*(pops-1) # 1 projection for 2 pops, 2 projections for 3 pops, etc
return [best_nneurons, max_synapses, max_total_synapses]
pops=10
max_synapses = sweep(pops=pops)
print("Testing max size of {} populations of N LIF neurons, all to all connections (pop0 to pop1, pop1 to pop2, etc.)".format(pops))
print("Max pre neurons: {}".format(max_synapses[0]))
print("Max synapses between 2 pops: {}".format(max_synapses[1]))
print("Max synapses total network : {}".format(max_synapses[2]))When you run this, notice that:
- The number of PEs used depends on the number of populations created.
- These populations are connected with all-to-all synapses, which scales very rapidly. The maximum number of synapses that will fit if a population is restricted to 1 core is around 10-20k.
- You can change the number of neurons of a population that are inside a core by uncommenting the
pop.set_max_atoms_per_core(50)line. There is a tradeoff between the number of cores/PEs used for the simulation, and the maximum number of synapses that can be mapped. - From these results, it is better to run simulations with sparse connections, or islands of densely connected populations connected to one another sparsely. Large dense networks are also possible, but with tradeoffs with regard to the number of cores utilized (and related increased communication times, power consumption, etc.)
Dry Run and other interesting options
You can do a dry run without submitting a simulation to the board by using the mapping_only option:
hw = hardware.SpiNNaker2Chip() # Notice: No board IP required.
hw.run(net, timesteps, mapping_only=True)This still lets you test the correctness of your code and even some py-spinnaker routines. See for example the code in the snn_profiling folder in py-spinnaker2 examples folder. Other interesting options of the hardware object:
- mapping_only (bool): run only mapping and no experiment on hardware. default: False
- debug(bool): read and print debug information of PEs. default: True
- iterative_mapping(bool): use a greedy iterative mapping to automatically solve MemoryError (maximizing number of neurons per core). default: FalseCurrently Known Problems
If you can’t seem to import the py-spinnaker2 library in Python, it is possible that your environment is using the wrong Python interpreter. Make sure that your PATH variable is not pointing to some paths outside of the container (for example, some conda installation in your home folder).
External Documentation
Documentation directly from SpiNNcloud:
Current limitations
At the moment, only the Python library py-spinnaker2 is available. In the future users should also gain access to the lower level C code defining the various neurons, synapses, and other simulation elements, so they can create custom code to be run on SpiNNaker.
There are presently some software based limitations to the number of SpiNNaker cores that can be used in a simulation, the possibility of setting up dynamic connections in a network, and possibly others. These will become available as the software for SpiNNaker is further developed. Keep an eye out on updates to the code repositories.