Data Pool
The GWDG data pool consists of datasets that are relevant to different user groups over a longer period of time. This includes datasets that are to be shared within a working group or with external parties. The data pool concept is a well-known and simple strategy for organising the sharing of curated datasets. A good example is the system implemented by DKRZ1.
This includes, for example:
- training data sets for machine learning applications
- open data sets of (inter-)governmental organizations
- open data sets of any HPC users
- project data for other projects to use
- semi-public project data that should be only shared upon application
- And many more!
Usage
Each data pool has a name, a version, content files (data and code), metadata files, and a README.md for other users to get started with the dataset. Pool data is either public (everyone on the cluster can access them) or non-public (grant access to specific other projects and users), but pool metadata and the README.md are always public. A website listing all available data pools is planned.
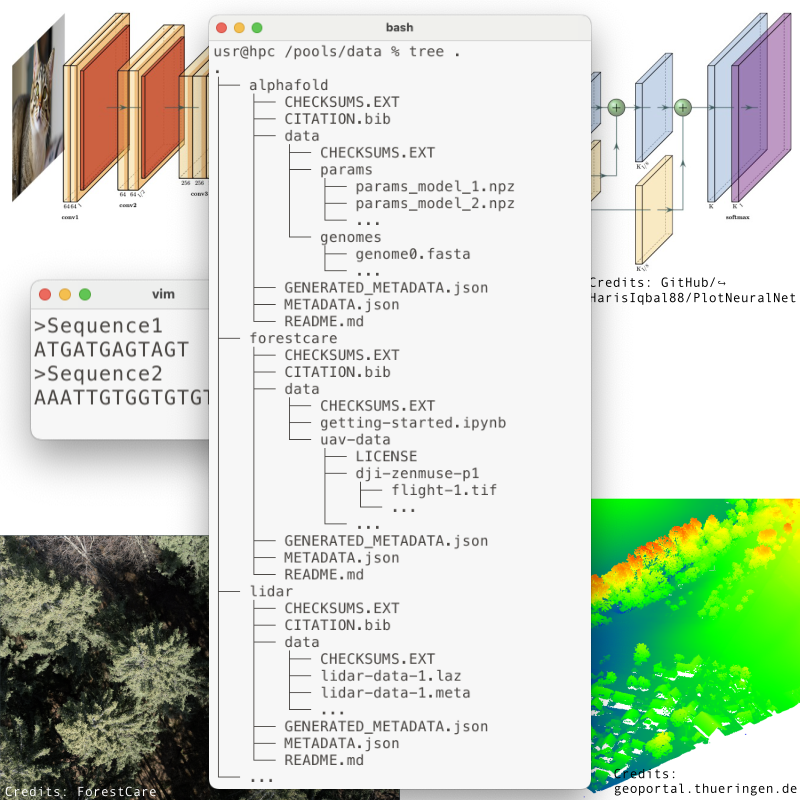
All datasets are centrally located in the /pools/data. The path to each pool follows the scheme
/pools/data/PROJECT/POOLNAME/POOLVERSIONwhere PROJECT is the project’s HPC Project ID (see Project Structure), POOLNAME is the name of the pool, and POOLVERSION is the specific version of the pool.
The file structure inside each data pool is
| Path | Type | Description |
|---|---|---|
public | file | DRAFT ONLY: Optional empty file. If present, the pool will be public. |
README.md | file | Documentation for the pool |
METADATA.json | file | Pool metadata |
CITATION.bib | file | BibTeX file with references to cite if using everything in the pool |
GENERATED_METADATA.json | file | GENERATED: Pool metadata that can’t be generated before submission |
CHECKSUMS.EXT | file | GENERATED: Tagged checksums of content/CHECKSUMS_*.EXT and all top-level files ¹ |
.git* | dir/files | GENERATED: Git repo for the top-level files other than GENERATED_METADATA.json |
content/ | directory | Directory holding pool content (non-public for non-public pools) |
content/CHECKSUMS_code.EXT | file | GENERATED: Tagged checksums of every file in content/code ¹ |
content/CHECKSUMS_data.EXT | file | GENERATED: Tagged checksums of every file in content/data ¹ |
content/.git* | dir/files | GENERATED: Git repo for the content/code directory and content/CHECKSUM_*.EXT files |
content/code/ | directory | Directory holding pool data |
content/code/* | files/dirs | The actual code of the data pool |
content/data/ | directory | Directory holding pool data |
content/data/* | files/dirs | The actual data of the data pool |
[1]: EXT is an extension based on the checksum algorithm (e.g. sha256 for SHA2-256).
Creation
Pools have to go through several phases.

Data Pool Workflow
Overview of the data pool creation process.
0. Prerequisite: Registered project
Only projects in the HPC Project Portal (see Project Management) are eligible to create pools. See Getting An Account for information on how to apply for a project.
Warning
NHR/HLRN projects created before 2024/Q2 must migrate to the HPC Project Portal before being eligibile to create pools. See the NHR/HLRN Project Migration page for information on migration.
1. Requesting data pool staging area
A project’s draft pools are created in a staging area under /pools/data-pool-staging/PROJECT.
Initially, projects don’t have access to the staging area.
A project PI can request access to the staging area via a support request (see Start Here for the email address to use).
The request should include a rough estimate of how much disk space and how many files/directories will be used.
If approved, a directory in the staging area is created for the project.
Each individual draft pool in preparation should use a separate subdirectories of the project’s staging directory, specifically /pools/data-pool-staging/PROJECT/POOL/VERSION where POOL is the pool name and VERSION is its version.
Info
The maximum number of files/directories in a pool is limited in order to improve IO performance of anyone using the pool. For example, directories with a million files are not allowed because anyone using the pool would harm the performance of the filesystem for everyone. In many cases, it is possible to bundle together large sets of small files (see Reducing Filesystem Usage for tips).
2. Building pool draft
Project members setup draft pools in subdirectories of the staging directory (note that each individual pool is treated separately from this point in the workflow). The subdirectory must be POOL/VERSION so that the pool name and version can be deduced from the path. A template data pool is provided, which you can access via:
cp -r /pools/data-pool-template/* /pools/data-pool-staging/PROJECT/POOL/VERSION/The template contains the basic files that must be filled out for the pool and directories that must get files:
README.mdMETADATA.jsonCITATION.bibcontent/content/code/content/data/
Make sure to create an empty public file if you want the pool to be public (and make sure it doesn’t exist if the pool should not be public), which can be done by
touch /pools/data-pool-staging/PROJECT/POOL/VERSION/publicand
rm /pools/data-pool-staging/PROJECT/POOL/VERSION/publicPut the pool code and data into the content/code/ and content/data/ subdirectories respectively, whether copying it from elsewhere on the cluster or uploading it to the cluster (more details in our documentation on data transfer).
There are a few hard restrictions:
- No files or additional directories in
content/. Everything must go under thecontent/code/andcontent/data/subdirectories. - All symlinks must be relative links that stay entirely inside the
content/directory and must eventually terminate on a file/directory (no circular links) - File, directory, and symlink names must all meet the following requirements:
- Hard requirements:
- Must be UTF-8 encoded (ASCII is a subset of UTF-8)
- Must not contain newline characters (wrecks havoc on unix shells)
- Must be composed entirely of printable characters (only allowed whitespace is the space character)
- Must not be a single
-or double dash--(wrecks havoc on passing to command line utilities) - Must not be a tilde
~(wrecks havoc on unix shells) - Must not be
.git(git repos are forbidden in submitted pools so that the top-level and content git repos can work)
- Recommendations
- Do not start with a dash
-(wrecks havoc on passing to command line utilities) - Do not start with a dot
.(pools shouldn’t have hidden files, directories, and/or symlinks) - Do not start with
.git(could cause problems for the content git repo) - Do not include autosave and backup files since they just waste space (files ending in
~,.asv,.backup,.bak, and.old) - Minimize binary files under
content/codesince the content git repo would include them (such files almost always belong undercontent/data)
- Do not start with a dash
- Hard requirements:
You should use the CLI pool validator at /pools/data-pool-tools/bin/data-pool-tools-validate to validate the various parts of your draft pools like
/pools/data-pool-tools/bin/data-pool-tools-validate [OPTIONS] PATHwhere PATH is the part of your draft pool you want to validate or even the whole draft pool if you give the path to its directory.
The validator autodetects what is being validated based on the specific PATH.
See Validator for more information.
For a straightforward setup of the data pool, we will eventually provide tools (CLI / WebUI / …) to help you prepare the various files in your draft pool and to check the draft pool for problems.
3. Submitting pool for review
A project PI submits the draft pool to become an actual pool by creating a support request (see Start Here for the email address to use). The following additional information must be included in the support request
- Project ID (if the project’s POSIX group is
HPC_foo, then the Project ID isfoo) - Pool name
- Pool version
- What sorts of other projects would be interested in using the data.
The pool’s path must then be /pools/data-pool-staging/PROJECT/POOL/VERSION.
Eventually, this will be replaced with a web form.
Once the submission is received, a read-only snapshot of the draft pool will be created and the various generated metadata files (checksum files and GENERATED_METADATA.json) generated for review. The validator is run. If it fails, the submitter is notified and the read-only snapshot is deleted so that they can fix the problems and resubmit. If the validator passes, the submitter is notified and the draft pool goes through the review process:
- All other PIs of the project are notified with the location of the draft pool snapshot and instructions on how to approve or reject the pool.
- If all other PIs have approved the pool, the draft pool goes to the Data Pool Approval Team.
- If the Data Pool Approval Team approves, the pool is accepted. Otherwise, they will contact the PIs.
4. Publishing Pool
Once a pool has been fully approved, it will be published in the following steps:
- The pool is copied to its final location and permissions configured.
- The pool metadata is added to the pool index within the data catalogue to appear in our Data Lake
- The draft pool’s read-only snapshot is deleted.
Finally, your data pool is available on our HPC system to all users with a high-speed connection. Anyone can access the data directly using the path to your data pool /pools/data/PROJECT/POOL/VERSION.
5. Editing Pool
Projects are allowed to edit a pool, either submitting a new version, a non-destructive revision, or a correction. Non-destructive revisions allow the following:
- Changes to top-level metadata files when the history of old versions can be kept
- Changes to
content/code/*when the history of old versions can be kept - Adding new files, directories, and/or symlinks under
content/data
A correction is when a top-level metadata file or file under content/code must be changed and the history of the old versions destroyed and/or when existing files, directories, and/or symlinks under content/data must be changed, removed, or renamed.
These situations should ideally not happen, but sometimes they are necessary.
For example, one could be including an external image that seemed to be CC-BY (thus one can share it) but it turns that the person who claimed to be its owner actually stole it and the original owner does not license it as CC-BY and won’t allow it to be shared, and thus the file must be deleted outright and its content expunged (but not necessarily history of its existence).
If you need to edit your data pool, copy it to the staging directory and follow the process from step 3 Submitting pool for review except that the following additional pieces of information must be given
- The pool to be editted must be specified.
- It must be specified whether this is a new version, a non-destructive revision, or a correction for an existing version.
- Specify what is changed (e.g. changelog)
- If doing a non-destructive revision or a correction, explain why. This is particularly critical for corections since they are destructive operations which undermines the reproducibility of the scientific results others derive from the pool. These changes to a data pool version can mean that the citation that others used to acknowledge the usage of your provided data is technically not correct anymore.
6. Regular review
All data pools are periodically reviewed to determine whether the data pool should be retained or deleted (or optionally archived) when the requested availability window expires.
Managing Access to Non-Public Pools
For pools with non-public data, access to files under /pools/data/PROJECT/POOL/VERSION/content is restricted via ACL.
Read access is granted to all members of the project, and any additional projects (must be in the HPC Project Portal) or specific project-specific usernames that a PI specifies.
Initially, changing who else is granted access requires creating a support request.
Eventually, this will be incorporated directly into the HPC Project Portal.
Data Documentation and Reuse License/s
It is recommended to follow domain specific best practices for data management, such as metadata files, file formats, etc.
While helpful, this is not enough by itself to make a dataset usable to other researchers.
To ensure a basic level of reusability, each data pool has README.md and METADATA.json files in their top-level directory containing a basic description of the dataset and how to use it.
These files are also critical for informing others which license/s apply to the data. All data must have a license, which should conform to international standards to facilitate re-use and ensure credit to the data creators2. Different files can have different licenses, but it must be made clear to users of the pool which license each file uses. Common licenses are:
- The various Creative Commons licenses for text and images
- The various licenses approved by OSI for source code
- CC0 for raw numerical data (not actually copyrightable in many legal jurisdictions, but this makes it so everyone has the same rights everywhere)
In addition, a CITATION.bib file is required for correct citation of the dataset when used by other HPC users.
This is a good place for pool authors to place the bibliographic information for the associated paper/s, thesis, or data set citation, as some journals like Nature provide.
This is for all things that would have to be cited if the whole data pool is used.
If some data requires only a subset of the citations, that would be a good thing to mention in the documentation (either the README.md or some other way in under the content/ directory).
The design of these files is strongly inspired by DKRZ1.
Warning
All information in this README.md and the METADATA.json file are publicly available, including the names and email addresses of the PI/s and creator/s of the data pool.
General Recommendations
- Follow good data organization, naming, and metadata practices in your field; taking inspiration from other fields if there is none or if they don’t cover your kind of data.
- Include minimal code examples to use the data. Jupyter notebooks, org-mode files, etc. are encouraged. Please place them in the
content/codedirectory if possible. - For source code, indicate its dependencies and which environment you have successfully run it in.
- Add metadata inside your data files if they support it (e.g. using Attributes in NetCDF and HDF5 files).
- Provide specifications and documentation for any custom formats you are using.
- Use established data file formats when possible, ideally ones that have multiple implementations and/or are well documented.
- Avoid patent encumbered formats and codecs when possible.
- Bundle up large numbers of small files into a fewer number of larger files.
- Compress the data when possible if it makes sense (e.g. use PNG or JPEG instead of BMP).
- Avoid spaces in filenames as much as possible (cause havoc for people’s shell scripts).
- Use UTF-8 encoding and Unix newlines when possible (note, some formats may dictate other ones and some languages require other encodings).
Files and Templates
Submitted: public
This file in a draft pool, if it exists, indicates that the pool is public. If it does not exist, the pool is restricted. The file must have a size of zero. The easiest way to create it is via
touch /pools/data-pool-staging/PROJECT/POOL/VERSION/publicNote
Note that the file is not copied to snapshots or the final published pool.
In snapshots and published pools, the information on whether it is public or not is instead in GENERATED_METADATA.json.
Submitted: README.md
The README.md should document the data and its use, so that any domain expert can use the data without contacting the project members.
It must be a Markdown document following the conventions of CommonMark plus GitHub Flavored Markdown (GFM) tables.
It must be UTF-8 encoded with Unix line endings.
The data pool TITLE on the first line must be entirely composed of printable ASCII characters.
The template README.md structure is
# TITLE
## owner / producer of the dataset
## data usage license
## content of the dataset
## data usage scenarios
## methods used for data creation
## issues
## volume of the dataset (and possible changes thereof)
## time horizon of the data set on /pool/dataSubmitted: METADATA.json
The metadata written by the pool submitters.
It must be a JSON file.
It must be UTF-8 encoded with Unix line endings.
Dates and times must be in UTC.
Dates must be in the "YYYY-MM-DD" format and times must be in "YYYY-MM-DDThh:mm:ssZ" format (where Z means UTC).
The file should be human readable (please use newlines and indentation).
It is a JSON dictionary of dictionaries.
At the top level are keys of the form "v_NUM" indicating a metadata version under which the metadata for that version of the metadata are placed.
This versioning allows the format to evolve while making it extremely clear how each field should be interpreted (using the version of the key) and allowing the file to contain more than one version at once for wider compatibility.
At any given time, the submission rules will dictate which version/s the submitters are required/allowed to use.
Many fields are based on the CF Conventions.
Most string fields must be composed entirely of printable characters with the only allowed whitespace characters being space and for some the Unix newline \n.
The template METADATA.json is
{
"v_1": {
"title": "TITLE",
"pi": ["Jane Doe"],
"pi_email": ["jane.doe@example.com"],
"creator": ["Jane Doe", "John Doe"],
"creator_email": ["jane.doe@example.com", "john.doe@example.com"],
"institution": ["Example Institute"],
"institution_address": ["Example Institute\nExample Straße 001\n00000 Example\nGermany"],
"source": "generated from experimental data",
"history": "2024-11-28 Created.\n2024-12-02 Fixed error in institution_address.",
"summary": "Data gathered from pulling numbers out of thin air.",
"comment": "Example data with no meaning. Never use.",
"keywords": ["forest-science", "geophysics"],
"licenses": ["CC0-1.0", "CC-BY-4.0"]
}
}Version 1
REQUIRED
The key is "v_1". The fields are
| Key | Value type | Description |
|---|---|---|
title | string | Title of the pool (must match TITLE in the README.md |
pi | list of string | Name/s of the principal investigator/s |
pi_email | list of string | Email address/es of the PI/s in the same order as "pi" - used for communication and requests |
creator | list of string | Name/s of the people who made the pool |
creator_email | list of string | Email address/es of the people who made the pool in the same order as "creator" |
institution | list of string | Names of the responsible institutions (mostly the institutions of the PI/s) |
institution_address | list of string | Postal/street address of each institution properly formatted with newlines (last line must be the country) in the same order as "institution". Must be sufficient for mail sent via Deutsch Post to arrive there. |
source | string | Method the data was produced (field from CF Conventions) |
history | string | Changelog style history of the data (will have newlines) (field from CF Conventions) |
summary | string | Summary/abstract for the data |
comment | string | Miscellaneous comments about the data |
keywords | list of string | List of keywords relevant to the data |
licenses | list of string | List of all licenses that apply to some part of the contents. Licenses on the SPDX License List must use the SPDX identifier. Other licenses must take the form "Other -- NAME" given some suitable name (should be explained in the documentation). |
Submitted: CITATION.bib
What paper(s), thesis(es), report(s), etc. should someone cite when using the full dataset? Written by the pool submitter. If it’s empty, the dataset cannot be cited in publications without contacting the author(s) (possibly because a publication using it hadn’t been published at the time of the pool submission).
Tip
If the citations required change for some reason (say, a manuscript is published and the citation should be changed from the preprint to the paper), you can submit a non-destructive revision for the pool version
It is a BibTeX file.
It must be ASCII encoded with Unix line endings.
The encoding is restricted to ASCII so it is compatible with normal BibTeX.
Otherwise, a user would have to use bibtex8, bibtexu, or BibLaTeX.
See https://www.bibtex.org/SpecialSymbols for how to put various non-ASCII characters into the file.
An example CITATION.bib would be
@Misc{your-key,
author = {Musterfrau, Erika
and Mustermann, Max},
title = {Your paper title},
year = {2024},
edition = {Version 1.0},
publisher = {Your Puplisher},
address = {G{\"o}ttingen},
keywords = {ai; llm; mlops; hpc },
abstract = {This is the abstract of your paper.},
doi = {10.48550/arXiv.2407.00110},
howpublished= {\url{https://doi.org/10.48550/arXiv.2407.00110},
}Generated: Top-Level Git Repo
Git repo generated during submission of the draft pool and not controlled by the submitters. It tracks the following files across revisions of the pool version:
README.mdMETADATA.jsonCITATION.bibCHECKSUMS.EXT(generated)
and the various Git support files (e.g. .gitignore).
Each revision of the pool version generates gets a new tag of the form rX where X is an incrementing base-10 number starting from 0 (the first revision is r0).
The latest revision is always checked out.
If you want to access an earlier revision, it is best to clone the repo and then checkout using the tag of the revision you want.
The list of revisions, their tags, and commit hashed can be found in the GENERATED_METADATA.json.
Generated: Content Git Repo
Git repo generated during submission of the draft pool and not controlled by the submitters.
It tracks all the content of the pool except for content/data.
Specifically, it tracks the following content across revisions of the pool version:
content/CHECKSUMS_code.EXT(generated)content/CHECKSUMS_data.EXT(generated)content/code/*
and the various Git support files (e.g. .gitignore).
This means that the list of files under content/data and their checksums is stored and tracked across revisions, but not the contents themselves.
Each revision of the pool version generates gets a new tag of the form rX where X is an incrementing base-10 number starting from 0 (the first revision is r0).
The latest revision is always checked out.
If you want to access an earlier revision, it is best to clone the repo and then checkout using the tag of the revision you want.
The list of revisions, their tags, and commit hashed can be found in the GENERATED_METADATA.json.
Generated: GENERATED_METADATA.json
Metadata generated during submission of the draft pool and not controlled by the submitters.
It is a JSON file.
It is UTF-8 encoded with Unix line endings.
Dates and times are in UTC.
Dates are in the "YYYY-MM-DD" format and times are in "YYYY-MM-DDThh:mm:ssZ" format (where Z means UTC).
It is a JSON dictionary of dictionaries.
At the top level are keys of the form "v_NUM" indicating a metadata version under which the metadata for that version of the metadata are placed.
This versioning allows the format to evolve while making it extremely clear how each field should be interpreted (using the version of the key) and allowing the file to contain more than one version at once for wider compatibility
An example GENERATED_METADATA.json is
{
"v_1": {
"public": true,
"project_id": "project123",
"pool_id": "cooldata",
"version" "0.39",
"submitter": "Mno Pqr",
"submitter_email": "pqr@uni.com",
"commit_date": "2024-12-03",
"commit_history": [
[
"r0",
"2024-12-03",
"5aaf25abb31252e846260ccf97cac5b412c1b1919376624dd9b1085e3bc0a385",
"756bfb08970e7c8c6137429e2a7cd6d44726be105a7472d75aff66780f706621"
]
]
}
}Version 1
The key is "v_1". The fields are
| Key | Value type | Description |
|---|---|---|
public | boolean | Whether the pool is public or not |
project_id | string | The HPC Project ID of the project |
pool_id | string | Pool name, which is the name of its subdirectory |
version | string | Pool version string |
submitter | string | Name of the user who submitted the pool |
submitter_email | string | Email address of the user who submitted the pool |
commit_date | string | Date (UTC) the pool was submitted/finalized |
commit_history | list of list | All previous commits as lists in order (more recent is last) of tag/revision name, "commit_date", commit hash of the top-level git repo, and commit hash of the content git repo. |
Generated: content/CHECKSUMS_code.EXT
Checksum file generated during submission containing the tagged checksums of all files under content/code.
The extension .EXT is based on the checksum algorithm (e.g. .sha256 for SHA-2-256).
Tagged checksums include the algorithm on each line and are created by passing the --tag option to Linux checksum programs like sha256sum.
If a data pool only has the data file content/code/foo, the content/CHECKSUMS_code.sha256 file would be something like
SHA256 (content/code/foo) = 7d865e959b2466918c9863afca942d0fb89d7c9ac0c99bafc3749504ded97730Generated: content/CHECKSUMS_data.EXT
Checksum file generated during submission containing the tagged checksums of all files under content/data.
The extension .EXT is based on the checksum algorithm (e.g. .sha256 for SHA-2-256).
Tagged checksums include the algorithm on each line and are created by passing the --tag option to Linux checksum programs like sha256sum.
If a data pool only has the data file content/data/foo, the content/CHECKSUMS_data.sha256 file would be something like
SHA256 (content/data/foo) = 7d865e959b2466918c9863afca942d0fb89d7c9ac0c99bafc3749504ded97730Generated: CHECKSUMS.EXT
Checksum file generated during submision containing the tagged checksums of the following files
README.mdMETADATA.jsonCITATION.bibcontent/CHECKSUMS_code.EXTcontent/CHECKSUMS_data.EXT
The extension .EXT is based on the checksum algorithm (e.g. .sha256 for SHA-2-256).
Tagged checksums include the algorithm on each line and are created by passing the --tag option to Linux checksum programs like sha256sum.
An example CHECKSUMS.sha256 file would be something like
SHA256 (README.md) = 7d865e959b2466918c9863afca942d0fb89d7c9ac0c99bafc3749504ded97730
SHA256 (METADATA.json) = d865e959b2466918c9863afca942d0fb89d7c9ac0c99bafc3749504ded977307
SHA256 (CITATION.bib) = 865e959b2466918c9863afca942d0fb89d7c9ac0c99bafc3749504ded977307d
SHA256 (content/CHECKSUMS_code.sha256) = 65e959b2466918c9863afca942d0fb89d7c9ac0c99bafc3749504ded977307d8
SHA256 (content/CHECKSUMS_data.sha256) = df2940bf16f5ada77b41f665e59e4433cff2c8ebc42e23f4b76e0187c187b73eGet in Contact With Us
If you have any questions left, that we couldn’t answer in this documentation, we are happy to get contacted by you via Ticket (E-Mail to our support addresses). Please indicate “HPC-Data Pools” in the subject, so your request reaches us quickly and without any detours.
https://docs.dkrz.de/doc/dataservices/finding_and_accessing_data/pool-data/index.html ↩︎ ↩︎
The PI of the data project is responsible to make sure, that this is in line with the respective data licence. ↩︎